
Level Up Your Training: Unleashing the power of serious games in your workplace
2023-06-29
How CRISPR Crunch transformed from Tower Defense to Pattern-Matching Puzzle
2023-10-12Building a "Puzzle Factory" for CRISPR Crunch
CRISPR Crunch is a fast-paced puzzle game for mobile, in which the player fends off waves after wave of attacking viruses. To kill a virus, you match the sequence of colors on the virus DNA with those in the hexagonal grid in the center of the screen.
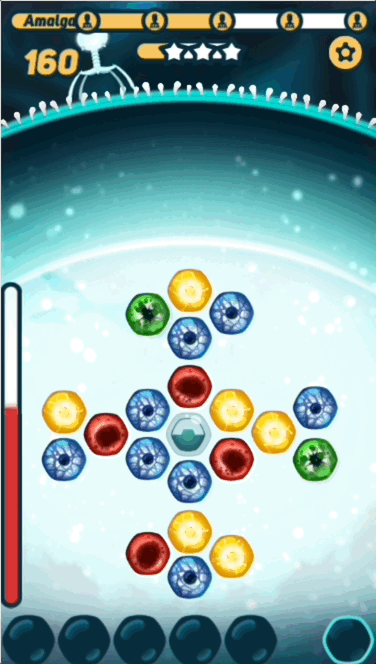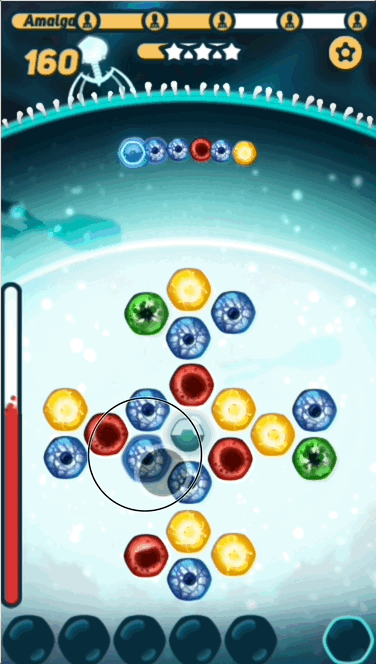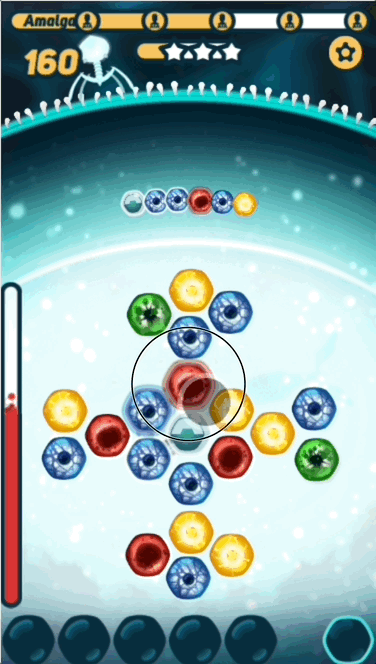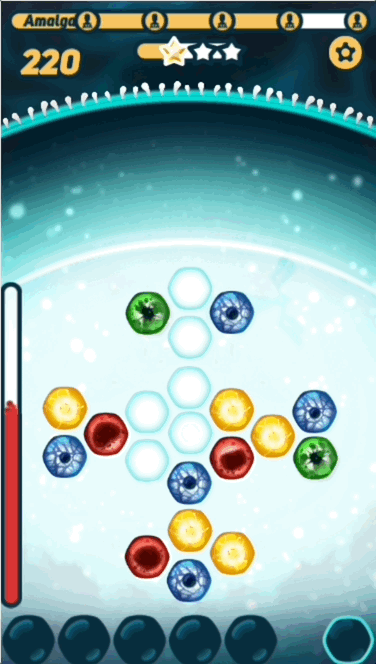
The basic gameplay is pretty simple - the player has to find the right path on the grid before the virus DNA reaches it, or they take damage. What makes the game interesting is challenging the player with sequences that are at the right level of difficulty- not too hard, and not too easy.
So how do we go about that? That’s the subject of this blog post.
Round 2, Fight!
In an earlier version of the game, we simply generated grids randomly. Then, we would pick a random sequence from the grid that the virus would inject.
This concept got put to the test when we got accepted to the Stunfest game festival in Rennes, France.
Given its roots in fighting game competitions, we suspected that Stunfest attendees would be excited to compete for the high score of the day, and we were right!
With just a simple giveaway, players came back again and again to reach amazing scores. It was a lot of fun to watch.
However, these players gave us feedback that we should focus on tuning the difficulty more precisely. Since our algorithm was simply picking random paths, sometimes it would pick a path that was really obvious, and other times one that was insanely tricky to find.
So on our way back from the festival, we decided to focus on finding a better way to generate paths of a given difficulty. But how?
Puzzle Creation
Before we dive in, let me explain how we named things, so the rest of this post makes more sense:
- Nucleotides - The "colored balls" that represent DNA bases
- Sequence - The nucleotides that the virus injects from the top of the screen. Those are the puzzles that the player is trying to solve
- Grid - The nucleotides in the hexagonal grid in the middle of the screen, where the player picks from
- Path - The series of nucleotides that the player picks, trying to solve the puzzle
- PAM - The first nucleotide in both sequences and paths, that looks a little bit like interlocking puzzle pieces
You might be surprised that I'm calling the sequence the puzzle, instead of the grid. This is because in CRISPR Crunch, the grid nucleotides that the player puts in their path are replaced after they are used. The result is that the grid nucleotides are constantly changing, so we can't "design" the grid to fit a given sequence.
Anyway, back to generating sequences. My first attempt was based on characterizing a path by how “rare” it was in the grid. My reasoning was that the more rare it is, the harder it is to find for the player.
For small grids, you can actually enumerate (list) all the paths that exist, and count up all the duplicates. So say in a given grid there are 10 occurences of yellow-yellow-yellow (YYY), but only 3 blue-blue-yellow (BBY). My reasoning would conclude that YYY is more than 3 times twice as hard to find as BBY, and therefore three times as difficult a puzzle.

Caption: I made this example by hand, I may have counted wrong ;)
In practice, enumerating large grids was taking too long on mobile (at least without multithreading, which I was trying to avoid), so I “sampled” paths in a Monte Carlo fasion, and put them in a list, ordered by how rare they were. I could then tell the algorithm to pick from a given part of the list to obtain the difficulty I was looking for.
Into the Player’s Mind
Although this first version matched my intuition, over time we discovered that some paths that were “rare” were still pretty easy to find. In fact, sometimes the rarity actually made it easier to find!
For example, imagine a grid with only 3 red nucleotides, and the algorithm happened to pick all 3 for the sequence. By definition, this is very rare (even unique), but the player’s eye would immediately recognize it.

Caption: There might only be 1 occurrence of RRR, but it’s not exactly hard to find!
So, rarity clearly wasn’t the attribute I was looking for. But following this line of thinking led me to develop a newer version of the algorithm, based on a naive model of how the player might go about finding a path on the grid.
I imagined that the player probably starts at the PAM, and then follows the colors in the sequence one by one. If they get stuck in a dead-end, they just start again, looking somewhere else.
If I simply counted the number of steps it takes the player to find the path, this should give an estimation of the “mental cost” of finding a path.
Here’s some pseudocode to formalize the idea. As before, we start by generating a set of random paths. Then, for each path, we follow these steps:
function estimateCost(sequence):
successfullAttempts := 0
repeat n times:
steps := 1
indexInSequence := 1
currentNucleotide := pickPam()
while indexInSequence < length(path):
// Try to find a neighbor of the correct color
currentNucleotide := getNeighbor(currentNucleotide, sequence[indexInSequence])
if isValid(currentNucleotide):
// Continue the path
indexInSequence++
steps++
else:
// Give up and start again
break
// We found a solution
successfullAttempts++
return steps / successfullAttempts
Of course, sometimes there are multiple PAMs or multiple nucleotides of the same color to pick from. In this case, I just pick randomly, assuming the player can’t do much better either.
Interpretation
Simply put, my improved algorithm counts how many nucleotides it takes this naive approach to find the solution. The lower the number, then the easier it is to find the path. Higher numbers indicate having to start over again and again, meaning harder challenges.
To go back to our test case from earlier, the RRR path that exists just once in the grid, attached to a singular PAM, would be followed systematically by the algorithm, and therefore have a lower score.
In contrast, the algorithm would penalize a path where there are multiple starting and “branching” points that the player could potentially go down, and give it a higher difficulty score. In fact, the highest difficulty would be given to paths that are “rabbit holes”, where the player follows what looks like a promising path into the very end, when in fact the last nucleotide isn’t there.
In practice, we’ve found this algorithm gives much more consistent results, and it even adapts nicely to sequences of different lengths - since longer sequences will have an extra “cost” associated with them.
However, the algorithm doesn’t take into account duplicate colors, which are easier for the player to spot. But maybe this isn't a problem after all. Perhaps it's a bit like a “gift” to the player- some sequences just end up being a bit easier.
Wrapping Up
So what’s my takeaway for future game projects?
From now on, I plan on asking myself “What would the player do?” when approaching these kinds of level or puzzle design questions. Although computers often work very differently than humans, There’s something elegant about imitating a human approach when trying to design puzzles for them.
I’d love to hear what you think of CRISPR Crunch. You can try it out for yourself for free on Android or iOS.




